Combined Quantum Mechanical/Molecular Mechanical Methods (QM/MM)
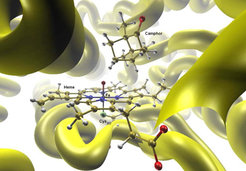
This research focused on hybrid approaches for large systems where the active center was treated by an appropriate quantum mechanical method, and the environment by a classical force field. It involved considerable method and code development. The QM/MM approach allowed a specific modeling of complex systems such that most of the computational effort was spent on the chemically important part. Following applications primarily addressed biocatalysis and aimed at a better understanding of enzymatic reactions including the role of the protein environment.
Methodological advances included:
- the definition of suitable QM/MM coupling schemes (embedding)
- the use of accurate correlated ab initio methods as QM component
- the use of polarizable Drude-type force fields as MM component
- the development of special techniques for QM/MM geometry optimizations
- the implementation of methods for QM/MM free-energy calculations
- the development of three-layer QM/MM/continuum treatments
- the extension of QM/MM methodology to electronically excited states
- the implementation of QM/MM-based quantum refinement for X-ray analysis
- the development of a modular QM/MM software environment (ChemShell)
While the QM/MM technology could be applied to many complex systems, the group was most interested in enzymatic reactions. Investigations at different QM/MM levels addressed biocatalysis by heme enzymes (e.g., cytochrome P450), molybdopterin enzymes (e.g., xanthine oxidase), cystein proteases, fluorinases, lipases, chorismate mutase, p-hydroxybenzoate hydroxylase, and cyclohexanone monooxygenase. In addition, the group also performed QM/MM studies on the spectroscopic properties of proteins, for examples on the Raman spectra of phycocyanin, the NMR spectra of vanadium-containing haloperoxidases, and the electronic spectra of fluorescent proteins. Surface hopping QM/MM simulations allowed to explore the excited-state dynamics of chromophores embedded in an environment.
